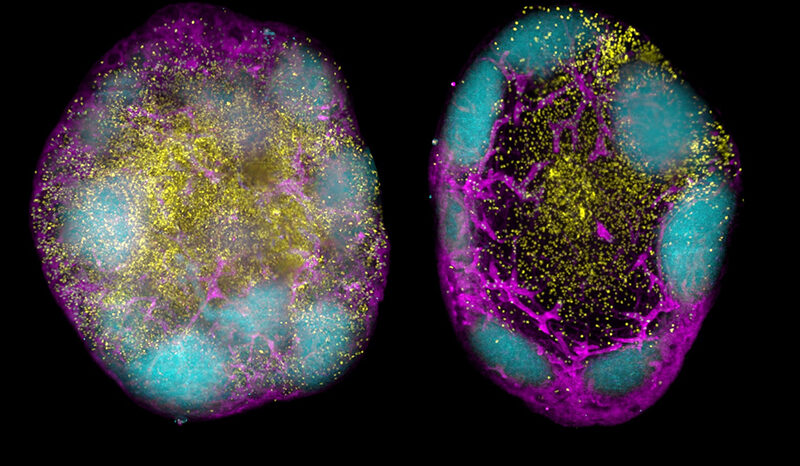
Broomfield BJ, Tan CW, Qin RZ, Abberger H, Duckworth BC, Alvarado C, Dalit L, Lee CL, Mugan RS, Mazrad ZAI, Muramatsu H, Mackiewicz L, Williams BE, Chen J, Takanashi A, Fabb S, Pellegrini M, Rogers KL, Moon WJ, Pouton CW, Davis MJ, Nutt SL, Pardi N, Wimmer VC, Groom JR. Transient inhibition of type I interferon enhances CD8+ T cell stemness and vaccine protection. Journal of Experimental Medicine. 2025;222(5):10.1084/jem.20241148
Broomfield BJ, Groom JR. Defining the niche for stem-like CD8+ T cell formation and function. Current Opinion in Immunology. 2024;89:10.1016/j.coi.2024.102454
Cooper L, Xu H, Polmear J, Kealy L, Szeto C, Pang ES, Gupta M, Kirn A, Taylor JJ, Jackson KJL, Broomfield BJ, Nguyen A, Gago da Graça C, La Gruta N, Utzschneider DT, Groom JR, Martelotto L, Parish IA, O’Keeffe M, Scharer CD, Gras S, Good-Jacobson KL. Type I interferons induce an epigenetically distinct memory B cell subset in chronic viral infection. Immunity. 2024;57(5):10.1016/j.immuni.2024.03.016
Abberger H, Groom JR. Macro-clusters: CD301b+ DCs prime Th2 responses. Journal of Experimental Medicine. 2024;221(4):10.1084/jem.20240088
Torres SV, Man K, Elmzzahi T, Malko D, Chisanga D, Liao Y, Prout M, Abbott CA, Tang A, Wu J, Becker M, Mason T, Haynes V, Tsui C, Shakiba MH, Hamada D, Britt K, Groom JR, McColl SR, Shi W, Watt MJ, Le Gros G, Pal B, Beyer M, Vasanthakumar A, Kallies A. Two regulatory T cell populations in the visceral adipose tissue shape systemic metabolism. Nature Immunology. 2024;25(3):10.1038/s41590-024-01753-9
Tellier J, Tarasova I, Nie J, Smillie CS, Fedele PL, Cao WHJ, Groom JR, Belz GT, Bhattacharya D, Smyth GK, Nutt SL. Unraveling the diversity and functions of tissue-resident plasma cells. Nature Immunology. 2024;25(2):10.1038/s41590-023-01712-w
Zaini A, Dalit L, Sheikh AA, Zhang Y, Thiele D, Runting J, Rodrigues G, Ng J, Bramhall M, Scheer S, Hailes L, Groom JR, Good-Jacobson KL, Zaph C. Heterogeneous Tfh cell populations that develop during enteric helminth infection predict the quality of type 2 protective response. Mucosal Immunology. 2023;16(5):10.1016/j.mucimm.2023.06.007
Wang N, Scott TA, Kupz A, Shreenivas MM, Peres NG, Hocking DM, Yang C, Jebeli L, Beattie L, Groom JR, Pierce TP, Wakim LM, Bedoui S, Strugnell RA. Vaccine-induced inflammation and inflammatory monocytes promote CD4+ T cell-dependent immunity against murine salmonellosis. PLOS Pathogens. 2023;19(9):10.1371/journal.ppat.1011666
Chan F, Coughlan HD, Ruhle M, Iannarella N, Alvarado C, Groom JR, Keenan CR, Kueh AJ, Wheatley AK, Smyth GK, Allan RS, Johanson TM. Survey of activation‐induced genome architecture reveals a novel enhancer of Myc. Immunology and Cell Biology. 2023;101(4):10.1111/imcb.12626
Groom JR, Vince JE. IL-1α-releasing TH17 cells live long and prosper. Nature Immunology. 2023;24(2):10.1038/s41590-022-01412-x